Illumina Cambridge Ltd v Latvia MGI Tech Sia
| Jurisdiction | England & Wales |
| Judge | Lord Justice Warby,Lord Justice Arnold,Lord Justice Nugee |
| Judgment Date | 17 December 2021 |
| Neutral Citation | [2021] EWCA Civ 1924 |
| Docket Number | Case No: A3/2021/0612 |
| Court | Court of Appeal (Civil Division) |
[2021] EWCA Civ 1924
Lord Justice Arnold
Lord Justice Nugee
and
Lord Justice Warby
Case No: A3/2021/0612
IN THE COURT OF APPEAL (CIVIL DIVISION)
ON APPEAL FROM THE HIGH COURT OF JUSTICE, BUSINESS AND PROPERTY
COURTS OF ENGLAND AND WALES,
INTELLECTUAL PROPERTY LIST, PATENTS COURT
Mr Justice Birss
Royal Courts of Justice
Strand, London, WC2A 2LL
Thomas Mitcheson QC, Thomas Hinchliffe QC, Miles Copeland, Isabel Jamal and Alice Hart (instructed by Allen & Overy LLP) for the Appellants
Iain Purvis QC, Piers Acland QC and Kathryn Pickard (instructed by Powell Gilbert LLP) for the Respondent
Hearing dates: 7–8 December 2021
Approved Judgment
This judgment was handed down remotely at 10.30am on 17 December 2021 by circulation to the parties or their representatives by email and by release to BAILII and the National Archives.
Introduction
This is an appeal by the Appellants (“MGI”) from an order of Birss J (as he then was) dated 18 February 2021 declaring, for the reasons given in his judgment dated 20 January 2021 [2021] EWHC 57 (Pat), that four patents owned by the Respondent (“Illumina”) were, as amended, valid and had been infringed by MGI. The judge also held that a fifth patent was invalid, but there is no appeal by Illumina against his order for revocation of that patent.
The four patents which are the subject of the appeal fall into two groups:
i) European Patent (UK) No. 1 530 578, European Patent (UK) No. 3 002 289 and European Patent (UK) No. 3 587 433 (“the Modified Nucleotide Patents”). These three patents are divisionals. The first two are entitled “Modified Nucleotides for Polynucleotide Sequencing” and the third is entitled “Modified Nucleotides”. They are based on an application filed on 22 August 2003. Although the earliest claimed priority is from a US application filed on 23 August 2002, Illumina only sought to maintain priority from the second priority document, a British application filed on 23 December 2002 (“P2”). It is common ground that the issues in relation to the Modified Nucleotide Patents may be determined by reference to the second of the three patents (“289”).
ii) EP (UK) No 2 021 415 entitled “Dye Compounds and the Use of Their Labelled Conjugates” (“the 415 Patent”). The application for this patent was filed on 16 May 2007 claiming priority from a US application filed on 18 May 2006.
The patents describe and claim inventions in the field of DNA sequencing, more specifically a technique known as “sequencing by synthesis”. The inventions described and claimed in the Modified Nucleotide Patents are concerned with using an azidomethyl group (-CH 2N 3) as a reversible chain terminator in sequencing by synthesis. The 415 Patent describes and claims a class of molecules useful in sequencing by synthesis consisting of three moieties: a nucleotide, a particular cleavable linker and a particular fluorescent dye.
The judge was faced with a considerable number of issues. On the appeal, only three of those issues are still live, namely:
i) Are the Modified Nucleotide Patents obvious over S. Zavgorodny et al, “1- Alkythioalkylation of Nucleoside Hydroxyl Functions and Its Synthetic Applications: A New Versatile Method in Nucleoside Chemistry”, Tetrahedron Letters, 32 (15), 7593– 7596 (1991) (“Zavgorodny”)?
ii) Are the Modified Nucleotide Patents entitled to priority from P2? It is common ground that, if the Modified Nucleotide Patents are not entitled to priority from P2, then they are obvious over Illumina's US Patent Application No. 2003/0104437 (“Barnes”), which was published on 5 June 2003.
iii) Is the 415 Patent obvious as a collocation of non-inventive features?
The Modified Nucleotide Patents
The skilled team
There was a substantial dispute before the judge as to the identity and attributes of the person or team skilled in the art to whom the Modified Nucleotide Patents were addressed. The judge found at [96] and [98] that they were addressed to a team working on research into sequencing by synthesis with two members. One member would have a background in molecular biology or genetics, with a focus on DNA sequencing in particular, the other member would have a background in organic chemistry. They would both have a post-graduate degree, probably a PhD but perhaps a Masters, and some years' research experience.
Common general knowledge
There was also a substantial dispute as to the skilled team's common general knowledge. Before turning to the judge's findings, it may assist comprehension if I give a couple of paragraphs of explanation based on the primer agreed between the parties for trial.
The structure of DNA. DNA consists of two complementary strands that wind around one another to form a double helix. The strands of DNA are made up of a string of individual nucleotides, which are composed of deoxyribose (a 5-carbon sugar), a nitrogenous base, and a phosphate group, as shown below. There are four different nucleotides in DNA, which differ from each other by their nitrogenous bases: adenine (A), cytosine (C), guanine (G), and thymine (T). Adenine and guanine are “purine” bases, whereas cytosine and thymine are “pyrimidine” bases.
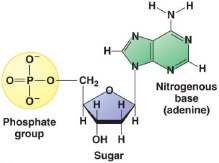
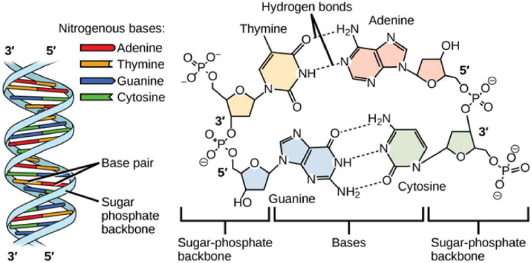
The nucleotides in each strand are linked by their phosphate group, which connects the 5' carbon atom (the atom in the -CH 2- group external to the ring) of one deoxyribose to the 3' carbon atom (the atom with a hydroxyl (-OH) substituent) of the deoxyribose of the next nucleotide, to form a sugar-phosphate backbone as shown below. The two complementary strands assemble together by pairing of the nitrogenous bases mediated by complementary hydrogen bonds, such that C pairs with G and A pairs with T.
Sanger sequencing. In the 1970s two ways of sequencing DNA were devised, each named after their inventors: Maxam-Gilbert sequencing and Sanger sequencing. Maxam-Gilbert sequencing was based on cutting the DNA strands using reagents which break the sequence at known places and analysing the results to deduce the original sequence. Sanger sequencing supplanted Maxam-Gilbert sequencing, and automated machines running Sanger sequencing were used in the human genome project in the 1990s.
The judge explained Sanger sequencing as follows:
“40. … Starting from the double stranded DNA of interest, a single strand is taken and used as a template in the method. DNA polymerase is used to add complementary nucleotide bases to the single template strand, one at a time. The complementarity of DNA means that the particular nucleotide base added at a given stage by the polymerase enzyme will be determined by the template sequence. So if the relevant nucleotide in the template is G then a C will be added to the growing complementary strand …. The trick to Sanger sequencing is that the free nucleotides to be added are not in their natural form.
41. Natural nucleotides have the capacity to form chains by joining together. … The chemical group at the 3' position is a hydroxyl (OH) group and the group at the 5' position is a triphosphate ester. The two ends connect together to form a link in the chain called a phosphodiester bond, liberating a molecule called pyrophosphate. A single strand of DNA therefore consists of a chain of these nucleotides and will have a ‘3’ end' at one end of the chain and a ‘5’ end' at the other end of the chain.
42. The way natural DNA synthesis works is that when a new nucleotide is added to the complementary strand, its 5' end is linked to the 3' end of the existing nucleotide which was already present. Once incorporated the unused 3' end of that newly linked nucleotide is ready to connect to the next fresh nucleotide, and so the complementary chain will grow.
43. In Sanger sequencing the new nucleotides are not natural because their 3' ends lack the 3' hydroxyl group. In the relevant naming convention it is called a dideoxynucleotide triphosphate (ddNTP). ….
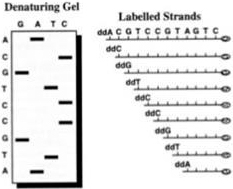
44. So in Sanger sequencing when a ddNTP is added, the chain cannot grow any further. Since there are four nucleotides (C, G, A and T) one can make four mixtures whereby each mixture has all four of C, G, A and T nucleotides in it but in each mixture, some examples of one kind of nucleotide are in the blocked ddNTP form instead of the natural dNTP form. Therefore when the DNA polymerase incorporates a ddNTP into the newly synthesised DNA strand, synthesis of the strand ceases (i.e. chain termination occurs). The Sanger sequencing process therefore results in the synthesis of a large number of copies of the template strand, which are terminated at random lengths according to the position at which a ddNTP is incorporated. A population of DNA strands of different lengths is therefore obtained, which end either with A, T, G, or C. These DNA strands of different lengths are then resolved using manual or automatic approaches and the DNA sequence can be understood. In a manual version of the process radiolabelled ddNTPs are used and the resulting radiolabelled copies of the template strand are size separated by gel electrophoresis. The DNA sequence of the template strand is determined from the order of the bands in the gel, as shown below:
”
Sequencing by synthesis. Sanger sequencing had certain drawbacks. The judge found that, by December 2002, there were a number of teams researching alternatives to Sanger sequencing. These included techniques which one would now call sequencing by synthesis. Only one such technique had been demonstrated to work, a technique called pyrosequencing. The details of pyrosequencing do not matter for present purposes. It suffices to say that it was different...
To continue reading
Request your trial-
Sandoz Ltd v Bristol-Myers Squibb Holdings Ireland Unlimited Company
...of this Court, namely FibroGen Inc v Akebia Therapeutics Inc [2021] EWCA Civ 1279 and Illumina Cambridge Ltd v Latvia MGI Tech SIA [2021] EWCA Civ 1924, it is not necessary to review those cases for present purposes. G 2/21 43 In G 2/21 the Enlarged Board of Appeal considered three questi......